Research Overview
My research focuses on unraveling the complex interactions among marine microbes across various trophic levels, and how these relationships are shaped by environmental factors and human activities. This work is crucial for understanding the fundamental processes that drive marine ecosystem dynamics and biogeochemical cycles.
Our research group employs a multidisciplinary approach, seamlessly integrating laboratory experiments with extensive field studies. This dual strategy allows us to investigate the physiology of key marine microbial groups under controlled conditions, explore ecological interactions and community dynamics in natural environments, trace the evolutionary pathways of marine microbes in response to environmental pressures, and quantify the impact of microbial activities on marine biogeochemistry.
By combining cutting-edge genomic techniques, advanced microscopy, biogeochemical analyses, and ecological modeling, we aim to build a comprehensive understanding of marine microbial ecosystems. Our work spans from the molecular level to ecosystem-wide processes, providing insights into microbial adaptation strategies to changing ocean conditions, the role of microbial interactions in shaping community structure and function, how anthropogenic stressors such as climate change and pollution alter microbial community dynamics, and the feedback mechanisms between microbial activities and global biogeochemical cycles.
Current Projects
Microbial Dynamics in Changing Seas
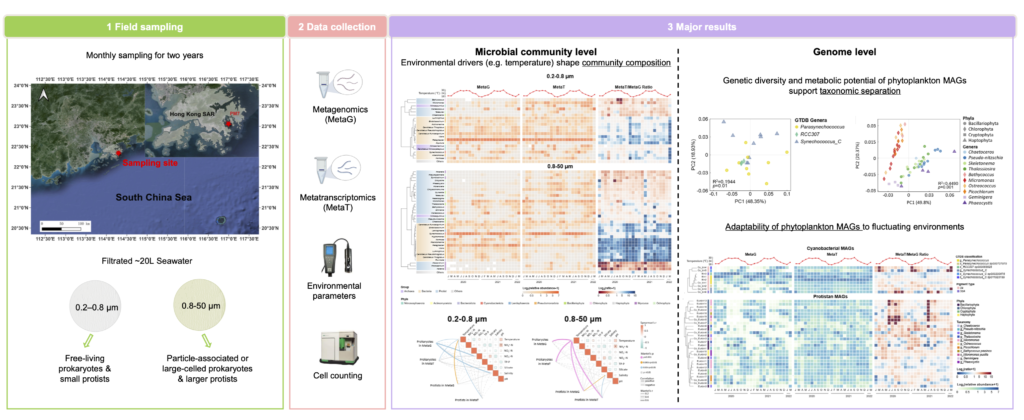
Our research explores the dynamic world of marine microbes across diverse ocean ecosystems using cutting-edge molecular techniques. We employ a comprehensive approach, including metagenomics, metatranscriptomics, long-read sequencing, Hi-C sequencing, and single-cell genomics, to study the entire spectrum of marine microbial life – from prokaryotes and eukaryotic microbes to viruses – and their complex interactions. Our work focuses on understanding microbial community changes as a whole, examining how these communities shift and adapt over time and across different ocean regions. We investigate both temporal and spatial dynamics, as well as the impact of anthropogenic activities on these microbial ecosystems. By integrating data from our advanced techniques, we’re creating a high-resolution, multi-dimensional picture of marine microbial communities. This research provides crucial insights into ocean food webs, global nutrient cycles, and the response of microbial communities to environmental changes and human influences.
Exploring algal ecology and genomics

We study the interactions between green algae and their associated viruses, with a particular emphasis on the Mamiellophyceae class.
This multifaceted study encompasses several key areas:
- Culturing and Isolation: We employ specialized techniques to culture Mamiellophyceae hosts and isolate their viruses from environmental samples. This process involves using selective media, flow cytometry, and plaque assays to obtain pure viral isolates, which are crucial for subsequent analyses.
- Viral Morphology: Using advanced microscopy techniques, we examine the structural characteristics of these viruses to understand their physical properties and infection mechanisms.
- Genomic Analysis: Through next-generation sequencing and bioinformatics, we are decoding the genetic makeup of these viruses, providing insights into their evolution and host-specificity.
- Infection Dynamics: By conducting time-course experiments, we are elucidating the complex processes of viral infection, replication, and host cell lysis. Furthermore, we are investigating how environmental factors such as temperature, light intensity, nutrient availability, and pH influence these infection dynamics.
- Global Distribution: Leveraging metagenomic and metatranscriptomic data from various oceanic regions, we are mapping the worldwide occurrence of these viruses to understand their ecological significance and biogeography.
A Holistic eDNA Approach to Marine Biodiversity in Hong Kong

Our research leverages environmental DNA (eDNA) techniques to revolutionize marine biodiversity monitoring in Hong Kong’s challenging aquatic environments. Traditional SCUBA-based visual surveys face significant limitations in these turbid waters, prompting us to develop a more efficient and accurate method. By analyzing eDNA from water and sediment samples, we aim to simultaneously quantify the diversity and abundance of coral and reef fish communities. Additionally, we are expanding our investigation to include 16S and 18S rRNA analyses, allowing us to characterize prokaryotic communities and eukaryotic phytoplankton. This comprehensive approach provides a holistic view of the local coral ecosystem, from microscopic organisms to larger reef inhabitants.
Previous Projects
Diatom-bacterium interactions
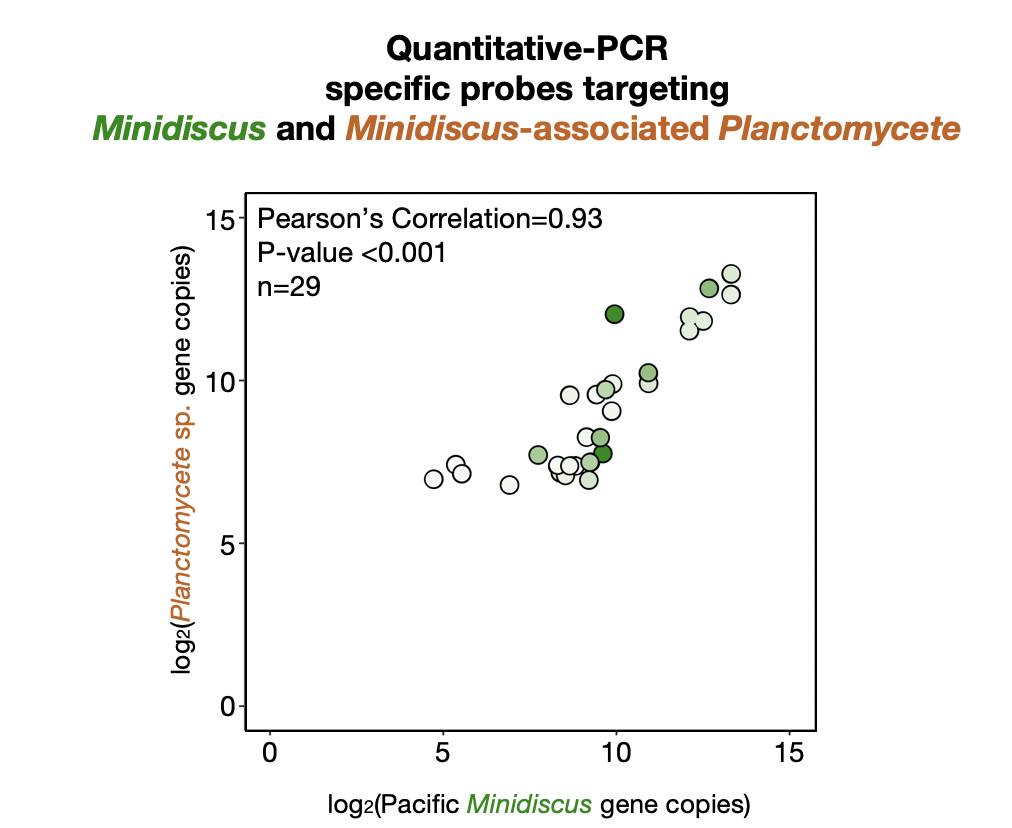
The co-occurrence of diatoms and bacteria over evolutionary time-scales appears to have dramatically influenced diatom gene content and physiology. Diatom-associated bacteria are thought to engage the host in a number of interactions, of both hostile and potentially mutualistic natures. Research to date has focused on individual cultured taxa or whole communities, and specific interactions between a diatom ‘host’ and its individual microbiome are difficult to discern in complex environmental systems. I use single-cell sorting coupled with genomic tools to identify bacteria that were physically-associated with diatoms. I examined diatom cells from a spring bloom in the northeastern Pacific Ocean. We found a highly divergent heterotrophic Planctomycete bacterium physically-associated with the diatom Minidiscus variabilis.
Fine-scale spatial microbial community studies
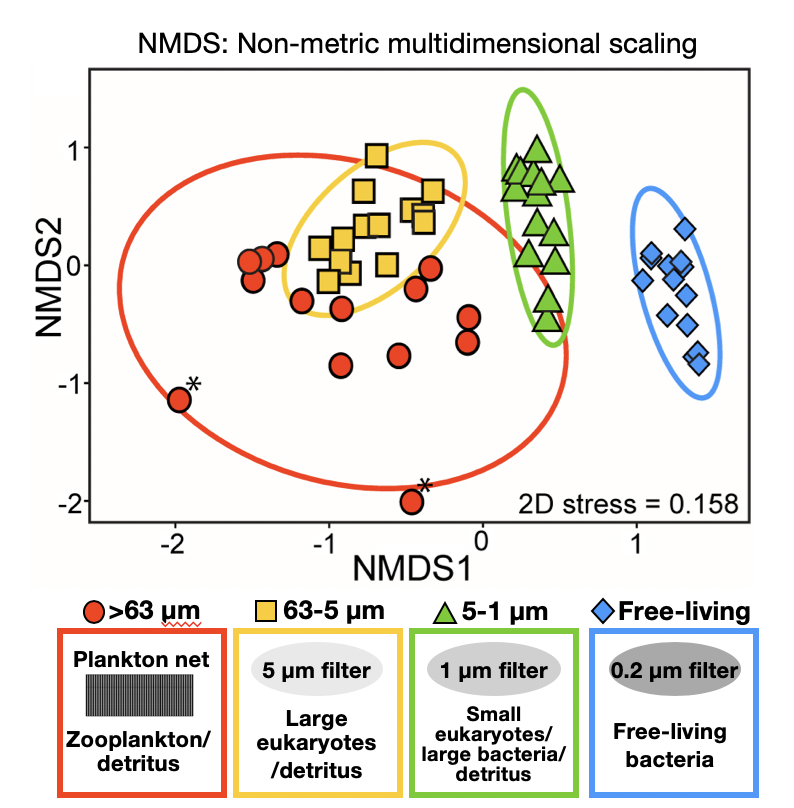
We investigated the temporal dynamics of particle-attached and free-living bacterial communities in coastal sea waters and found that microhabitats exert a stronger forcing on microbial communities than environmental variability.
Potential mechanisms of thermal adaptation in marine bacteria
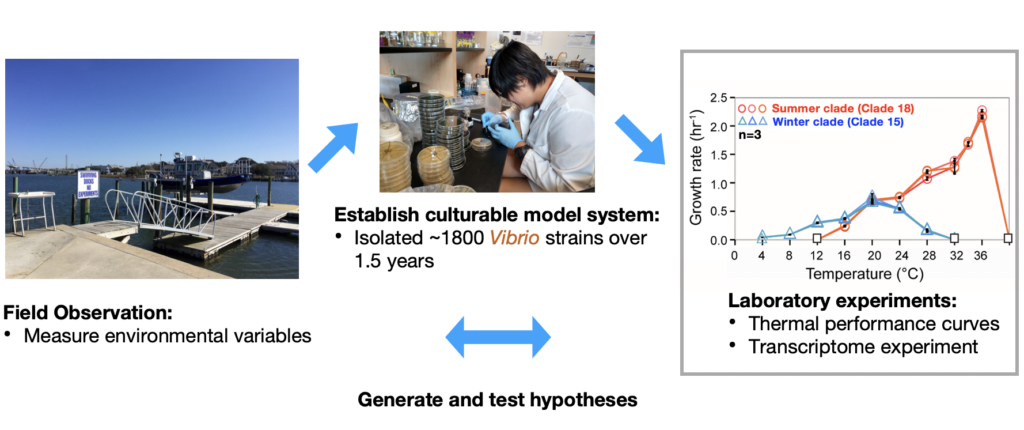
My early studies explored thermal adaptation in marine bacteria – Vibrios through a combination of strain-level population studies, growth rate experiments, genomics, transcriptomics, and fine-scale spatial community studies. I have found that closely-related vibrio strains observed under a range of temperature conditions exhibited thermal specialization at very fine-scale phylogenetic resolution. To follow up, I looked at how these closely-related strains regulate their genes to cope with different thermal stresses.
