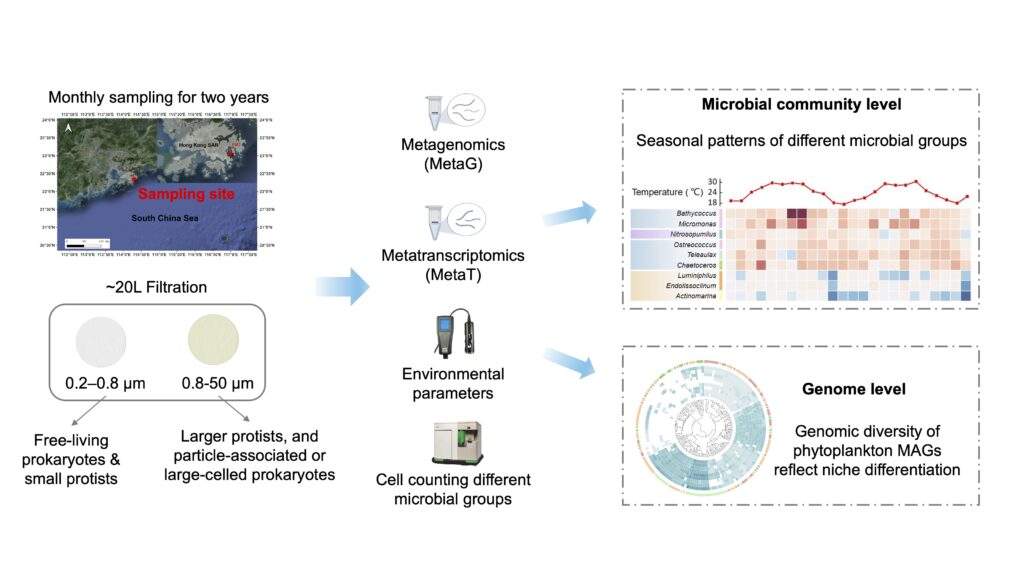
We’re proud to announce that Wenqian Xu, PhD candidate, from our group has published a paper in Science of the Total Environment. The study, “Revealing the intricate temporal dynamics and adaptive responses of prokaryotic and eukaryotic microbes in the coastal South China Sea,” offers new insights into marine microbial communities.
Wenqian’s two-year study combined metagenomics and metatranscriptomics to explore how microbial communities adapt to environmental changes. The research uncovered seasonal shifts in microbial composition and analyzed 37 phytoplankton metagenome-assembled genomes.
Congratulations to Wenqian and the team on this significant contribution to marine microbiology!